Artificial Intelligence in Analysis of Clinical Data

Artificial intelligence (AI) is wide-ranging branch of computer science concerned with building smart machines capable of performing tasks that typically require human intelligence. AI is an interdisciplinary science with multiple approaches, but advancements in machine learning and deep learning are creating a paradigm shift in virtually every sector of the tech industry.
The future of ‘standard’ medical practice might be here sooner than anticipated, where a patient could see a computer before seeing a doctor. Through advances in AI, it appears possible for the days of misdiagnosis and treating disease symptoms rather than their root cause to move behind us. Think about how many years of blood pressure measurements you have, or how much storage you would need to delete to fit a full 3D image of an organ on your laptop? The accumulating data generated in clinics and stored in electronic medical records through common tests and medical imaging allows for more applications of artificial intelligence and high-performance data-driven medicine. These applications have changed and will continue to change the way both doctors and researchers approach clinical problem-solving.
We used the latest algorithms and methods developed in AI to integrate experimentally observed high throughput data, such as genomes, transcriptomes, proteomes, metabolomes and metagenomes for development of efficient treatment strategies for metabolism related diseases.
Systems Biology in Integration of Clinical Data
 Systems biology is an integrative research strategy that combines experimental and computational biology to identify the molecular mechanisms underlying the components of a complex biological system and to obtain a quantitative description. This rapidly developing field integrates mathematical models with publicly available experimentally observed high throughput data, such as genomes, transcriptomes, proteomes, metabolomes and metagenomes, and reconstructs biochemical networks for Systematic analysis of complex systems using interdisciplinary computational tools. In the context of systems biology, the computational approach focuses on the dynamics and interplay between biological systems such as cells, tissues and organs using a holistic approach rather than reductionism that focuses on individual components and typically excludes information regarding time, space and context
Systems biology is an integrative research strategy that combines experimental and computational biology to identify the molecular mechanisms underlying the components of a complex biological system and to obtain a quantitative description. This rapidly developing field integrates mathematical models with publicly available experimentally observed high throughput data, such as genomes, transcriptomes, proteomes, metabolomes and metagenomes, and reconstructs biochemical networks for Systematic analysis of complex systems using interdisciplinary computational tools. In the context of systems biology, the computational approach focuses on the dynamics and interplay between biological systems such as cells, tissues and organs using a holistic approach rather than reductionism that focuses on individual components and typically excludes information regarding time, space and context
In systems biology, predictive mathematical models are employed for the analysis of given experimental data in a quantitative fashion, for gaining new biological knowledge or for performing predictive simulations. There are two approaches to systems biology: the top-down and the bottom-up approach. The top-down approach is a data-driven process
where high-throughput experimental data are analysed with the objective of finding patterns or the function of biological subsystems (or pathways) in the system being studied. By contrast, the bottom up approach is typically hypothesis driven where detailed knowledge of subsystems is reconstructed into a mathematical model that can be used to describe the whole system.
Integration of biological networks
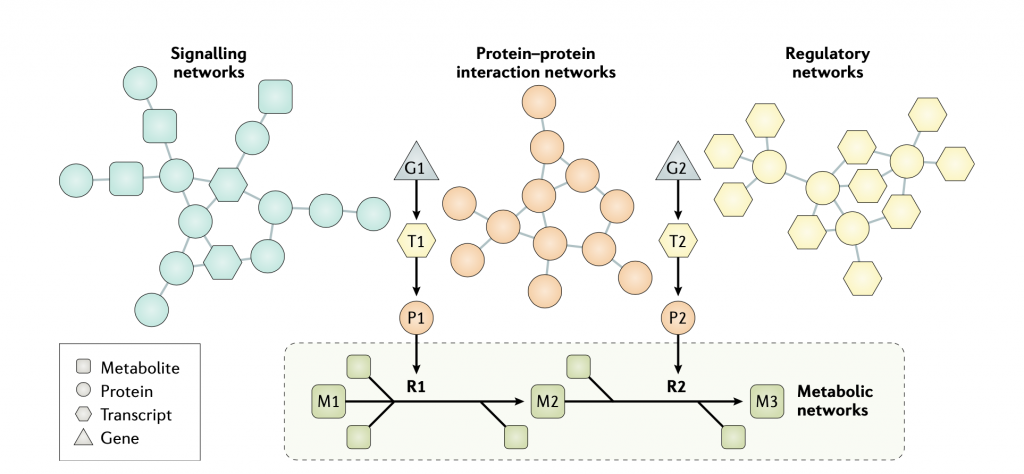 Individual biological networks, including metabolic, regulatory , signalling and protein–protein interaction networks, have been used in the analysis of omics data and to reveal the underlying molecular mechanisms of diseases to discover novel biomarkers and drug targets. High- quality omics data, including transcriptomics, proteomics, metabolomics and fluxomics, enable the generation of high- quality metabolic, signalling and protein–protein interaction networks. As shown in the figure, the effects of transcription factors and proteins on other proteins can be predicted by integration of different biological networks, and the level metabolites can be measured as a metabolic fingerprint in different clinical conditions. For example, gene 1 (G1) is transcribed to transcript 1 (T1), which is translated to protein 1 (P1); P1 then acts on metabolite 1 (M1) in reaction 1 (R1) to generate metabolite 2 (M2).
Individual biological networks, including metabolic, regulatory , signalling and protein–protein interaction networks, have been used in the analysis of omics data and to reveal the underlying molecular mechanisms of diseases to discover novel biomarkers and drug targets. High- quality omics data, including transcriptomics, proteomics, metabolomics and fluxomics, enable the generation of high- quality metabolic, signalling and protein–protein interaction networks. As shown in the figure, the effects of transcription factors and proteins on other proteins can be predicted by integration of different biological networks, and the level metabolites can be measured as a metabolic fingerprint in different clinical conditions. For example, gene 1 (G1) is transcribed to transcript 1 (T1), which is translated to protein 1 (P1); P1 then acts on metabolite 1 (M1) in reaction 1 (R1) to generate metabolite 2 (M2).
Integration of multiomics data using biological networks
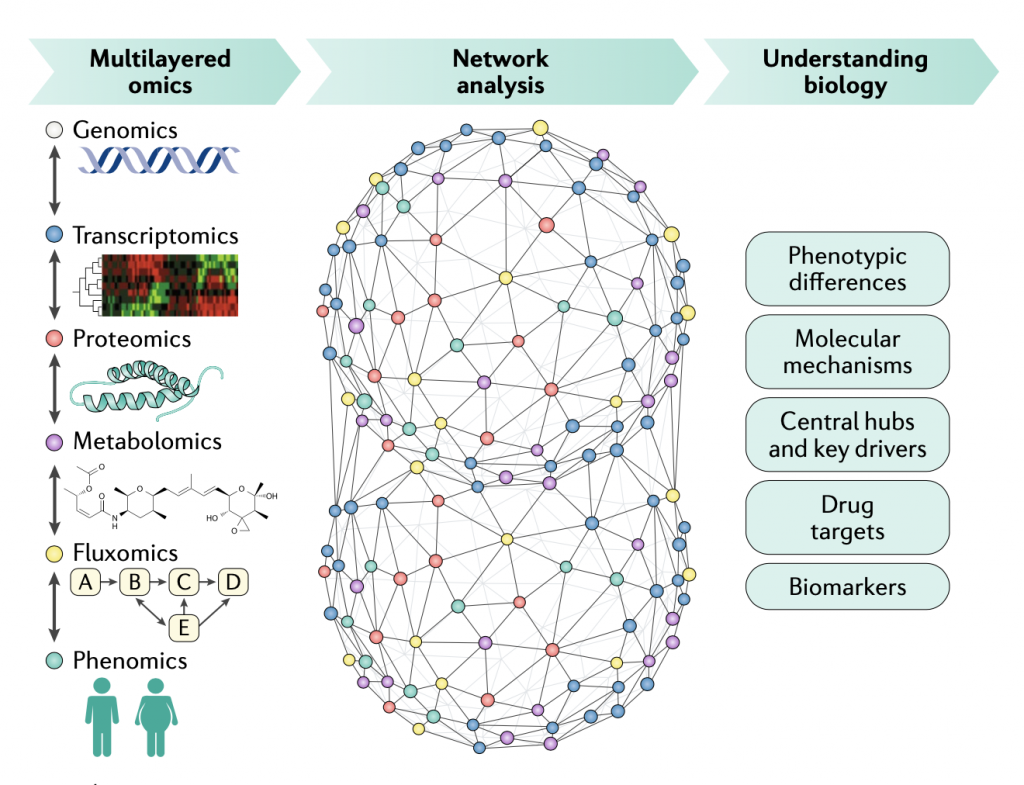 Technological advances in the generation of omics data have enabled the characterization of cells or tissues in health and disease states. However, no single omics data type is able to reveal the complex behaviour of cells or tissues in different states. In this context, biological networks can be used for integration of multiomics data. This approach might reveal phenotypic differences and underlying molecular mechanisms involved in the occurrence of the disease to ultimately identify novel biomarkers and drug targets. These integrative systems biology approaches can be used in the development of efficient treatment strategies and to achieve the goals of personalized and translational medicine.
Technological advances in the generation of omics data have enabled the characterization of cells or tissues in health and disease states. However, no single omics data type is able to reveal the complex behaviour of cells or tissues in different states. In this context, biological networks can be used for integration of multiomics data. This approach might reveal phenotypic differences and underlying molecular mechanisms involved in the occurrence of the disease to ultimately identify novel biomarkers and drug targets. These integrative systems biology approaches can be used in the development of efficient treatment strategies and to achieve the goals of personalized and translational medicine.
Metabolic alterations involved in the development of mitochondrial diseases
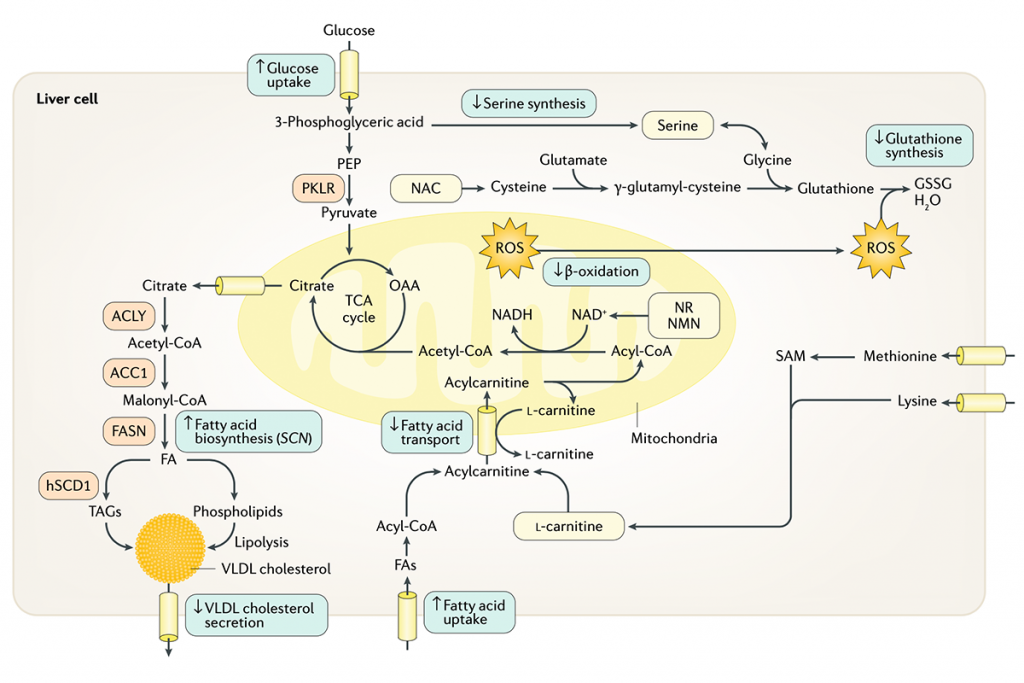 Liver Fat is caused by one or a combination of the following: improved delivery of fatty acids (FAs) to the liver ; increased endogenous FA synthesis; decreased mitochondrial FA oxidation; and deficient incorporation or export of triglycerides as VLDL cholesterol. New innovative approaches can be developed for effective treatment of liver fat by targeting one or more of these pathways. Proteins involved in de novo lipogenesis, including pyruvate kinase (PKLR), ATP- citrate synthase (ACLY), acetyl- CoA carboxylase 1 (ACC1), FA synthase (FASN) and stearoyl- CoA desaturase (hSCD1), have been targeted for inhibition of endogenous FA synthesis. Considering that altered NAD+ and glutathione metabolism (with an increased demand for NAD+ and glutathione) is a prevailing feature in NAFLD, supplementation of natural substances that might increase the level of NAD+ and glutathione could provide potential treatment strategies. For instance, natural substances, including serine, glycine and N- acetylcysteine (NAC), could be supplemented to boost the level of glutathione; the precursors of NAD+, including nicotinamide riboside (NR) and nicotinamide mononucleotide (NMN), could be supplemented to boost the level of NAD+; and L -carnitine could be supplemented to boost the transfer of FAs to the mitochondria. Supplementation of such natural substances (shown in yellow boxes) might eventually promote mitochondrial FA oxidation in liver, which can decrease the level of the accumulated fat in the liver. GSSG, glutathione (oxidized); OAA , oxaloacetate; PEP, phosphoenolpyruvate; ROS, reactive oxygen species; TAG, triacylglycerol; TCA , tricarboxylic acid; SAM, S- adenosyl-L- methionine.
Liver Fat is caused by one or a combination of the following: improved delivery of fatty acids (FAs) to the liver ; increased endogenous FA synthesis; decreased mitochondrial FA oxidation; and deficient incorporation or export of triglycerides as VLDL cholesterol. New innovative approaches can be developed for effective treatment of liver fat by targeting one or more of these pathways. Proteins involved in de novo lipogenesis, including pyruvate kinase (PKLR), ATP- citrate synthase (ACLY), acetyl- CoA carboxylase 1 (ACC1), FA synthase (FASN) and stearoyl- CoA desaturase (hSCD1), have been targeted for inhibition of endogenous FA synthesis. Considering that altered NAD+ and glutathione metabolism (with an increased demand for NAD+ and glutathione) is a prevailing feature in NAFLD, supplementation of natural substances that might increase the level of NAD+ and glutathione could provide potential treatment strategies. For instance, natural substances, including serine, glycine and N- acetylcysteine (NAC), could be supplemented to boost the level of glutathione; the precursors of NAD+, including nicotinamide riboside (NR) and nicotinamide mononucleotide (NMN), could be supplemented to boost the level of NAD+; and L -carnitine could be supplemented to boost the transfer of FAs to the mitochondria. Supplementation of such natural substances (shown in yellow boxes) might eventually promote mitochondrial FA oxidation in liver, which can decrease the level of the accumulated fat in the liver. GSSG, glutathione (oxidized); OAA , oxaloacetate; PEP, phosphoenolpyruvate; ROS, reactive oxygen species; TAG, triacylglycerol; TCA , tricarboxylic acid; SAM, S- adenosyl-L- methionine.
Host–gut microbiota crosstalk and effects on whole- body biological functions
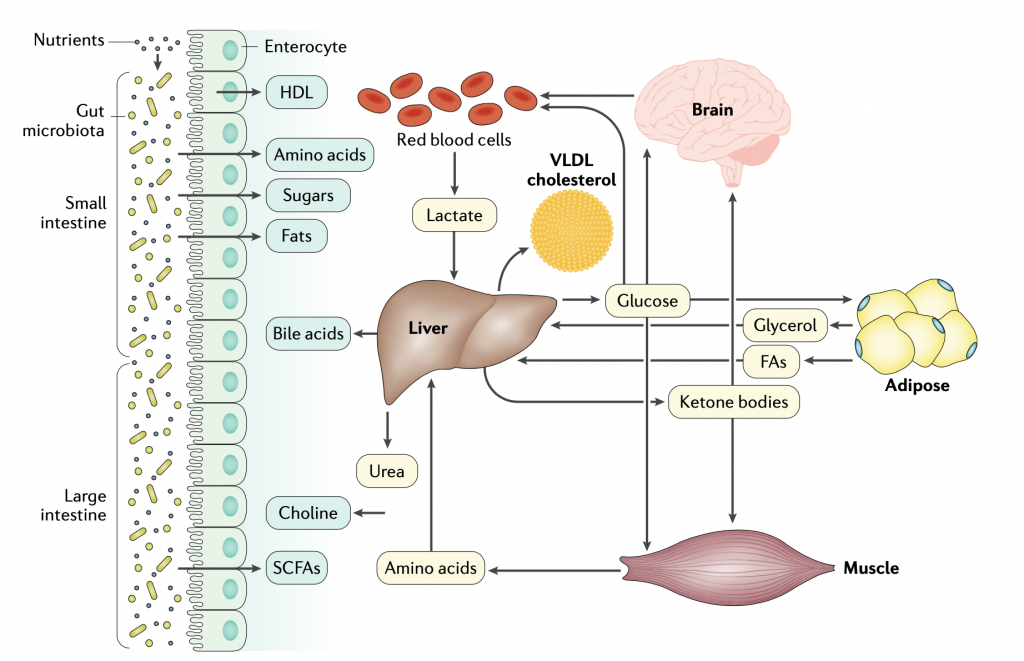 Understanding the interactions between different tissues and their interactions with the gut microbiota is necessary to gain detailed information about whole- body physiology. To study whole- body biological functions in health and disease states, simulation- ready cell specific or tissue- specific models, including those for the liver, adipose and muscle tissues, intestines, brain and red blood cells, as well as for the gut microbiota, can be integrated. Such knowledge can be used in elucidating the progression of metabolic diseases and for designing new therapies and clinical interventions. To study the crosstalk between liver and other human tissues, the secretion rate of VLDL cholesterol from liver can be measured, and these experimentally derived VLDL cholesterol kinetic data can be integrated with a liver genome- scale metabolic model (GSMM) and other constraints (glucose, ketone bodies, urea, bile acids and lactate) to simulate the dynamics of liver metabolism. The uptake and/or secretion rates of glycerol, fatty acids (FAs) and amino acids from adipose and muscle tissues can also be calculated on the basis of the body composition of each subject and integrated with the liver GSMM. The contribution of the gut microbiota to the host metabolism can also be predicted on the basis of diet, which accounts for the interactions among the microbiota. SCFAs, short- chain FAs.
Understanding the interactions between different tissues and their interactions with the gut microbiota is necessary to gain detailed information about whole- body physiology. To study whole- body biological functions in health and disease states, simulation- ready cell specific or tissue- specific models, including those for the liver, adipose and muscle tissues, intestines, brain and red blood cells, as well as for the gut microbiota, can be integrated. Such knowledge can be used in elucidating the progression of metabolic diseases and for designing new therapies and clinical interventions. To study the crosstalk between liver and other human tissues, the secretion rate of VLDL cholesterol from liver can be measured, and these experimentally derived VLDL cholesterol kinetic data can be integrated with a liver genome- scale metabolic model (GSMM) and other constraints (glucose, ketone bodies, urea, bile acids and lactate) to simulate the dynamics of liver metabolism. The uptake and/or secretion rates of glycerol, fatty acids (FAs) and amino acids from adipose and muscle tissues can also be calculated on the basis of the body composition of each subject and integrated with the liver GSMM. The contribution of the gut microbiota to the host metabolism can also be predicted on the basis of diet, which accounts for the interactions among the microbiota. SCFAs, short- chain FAs.
